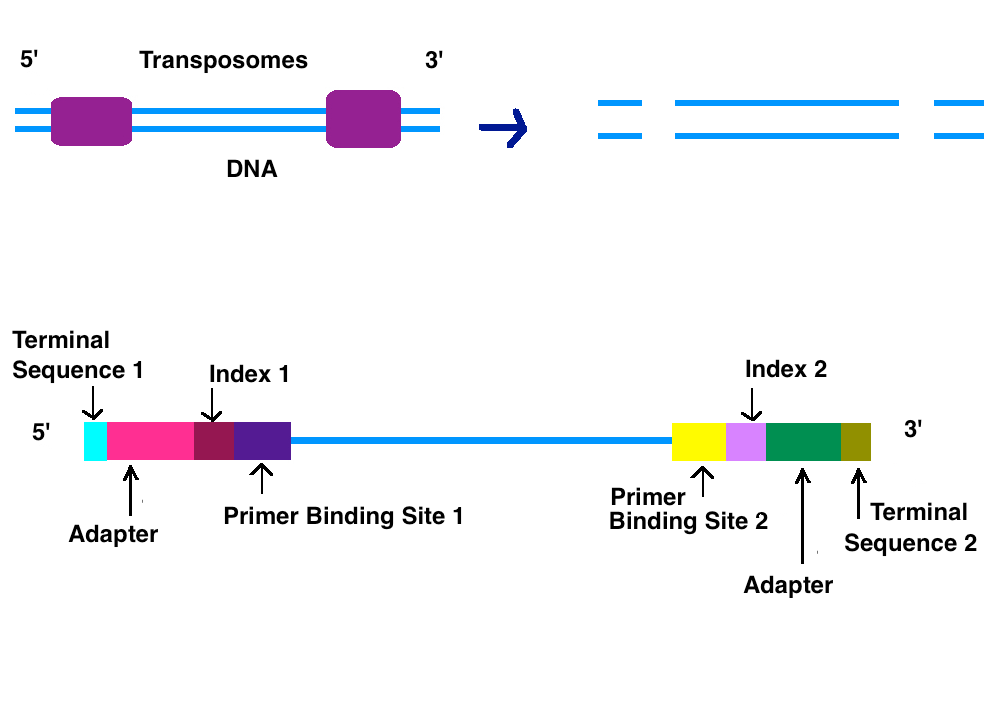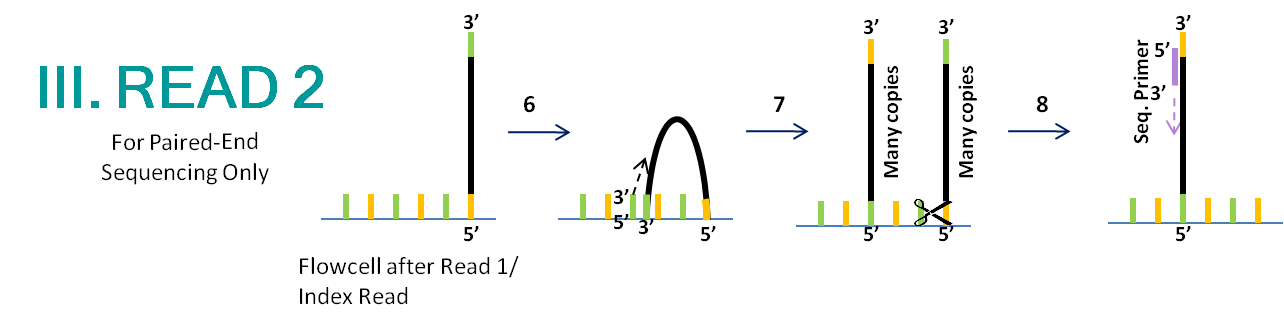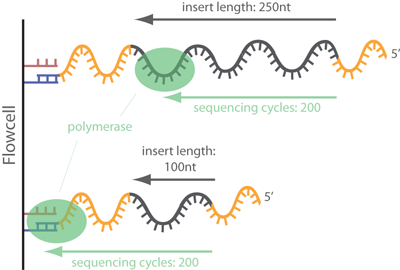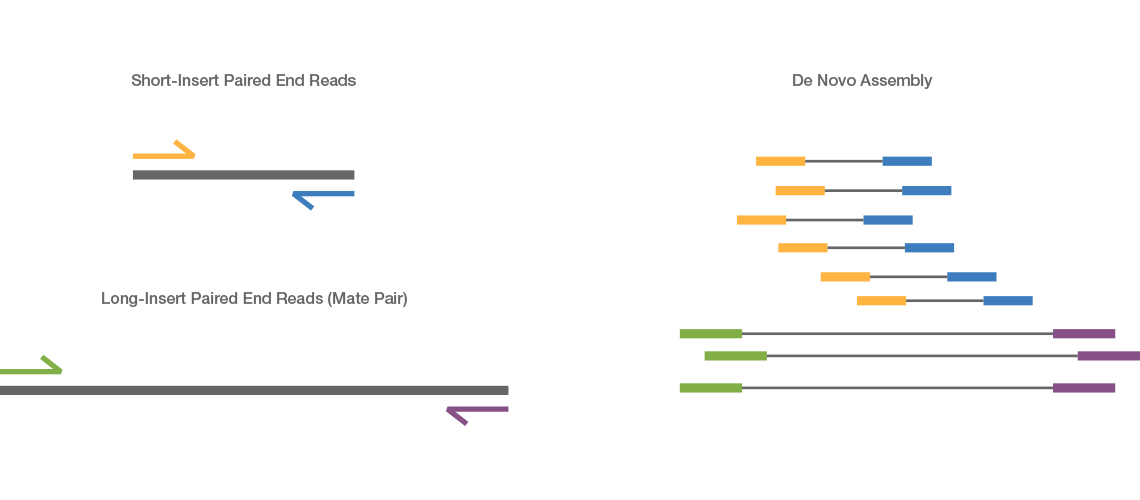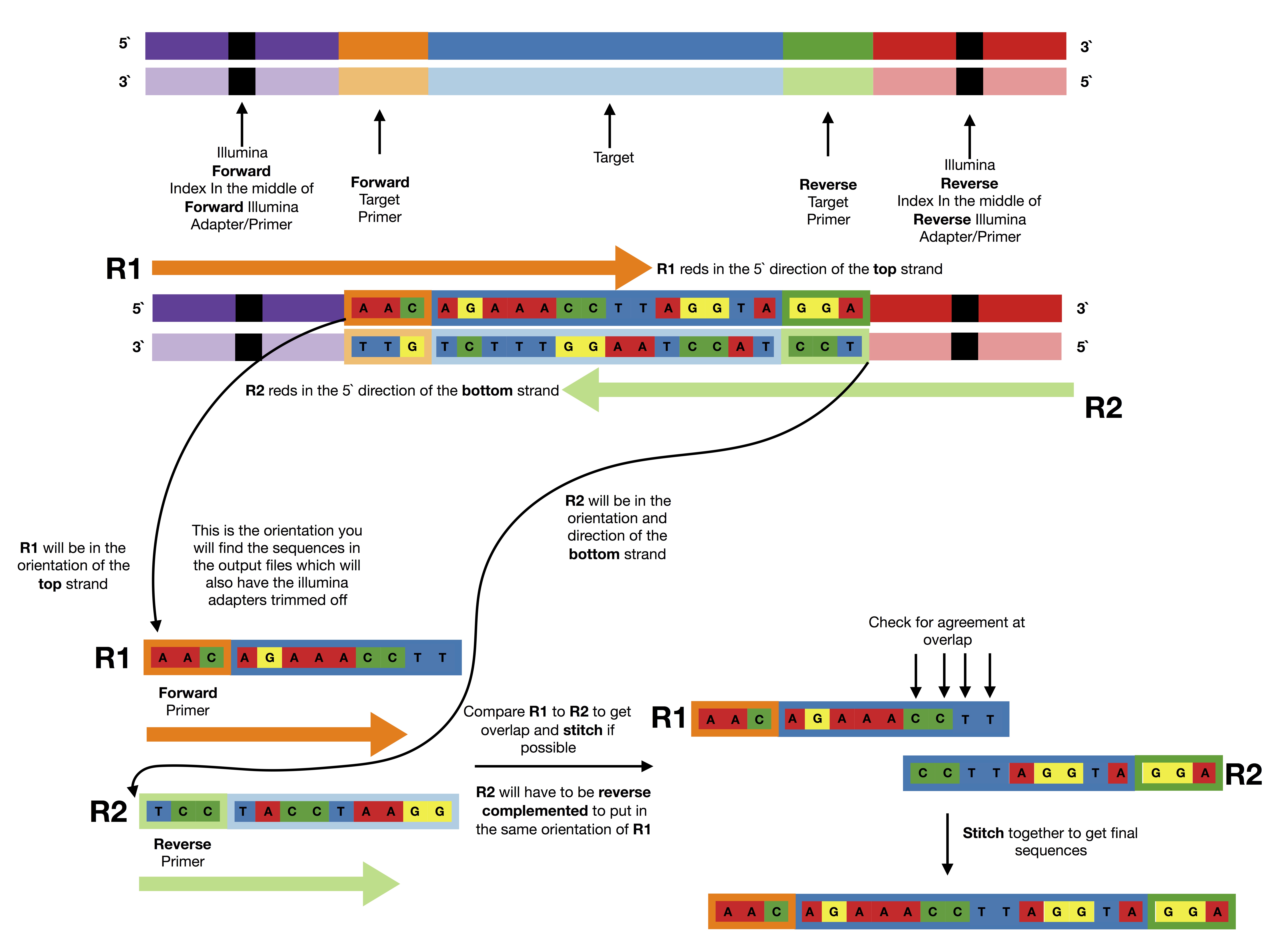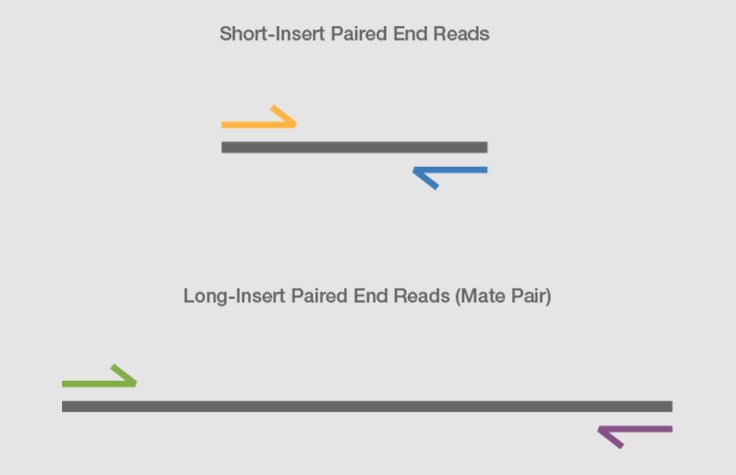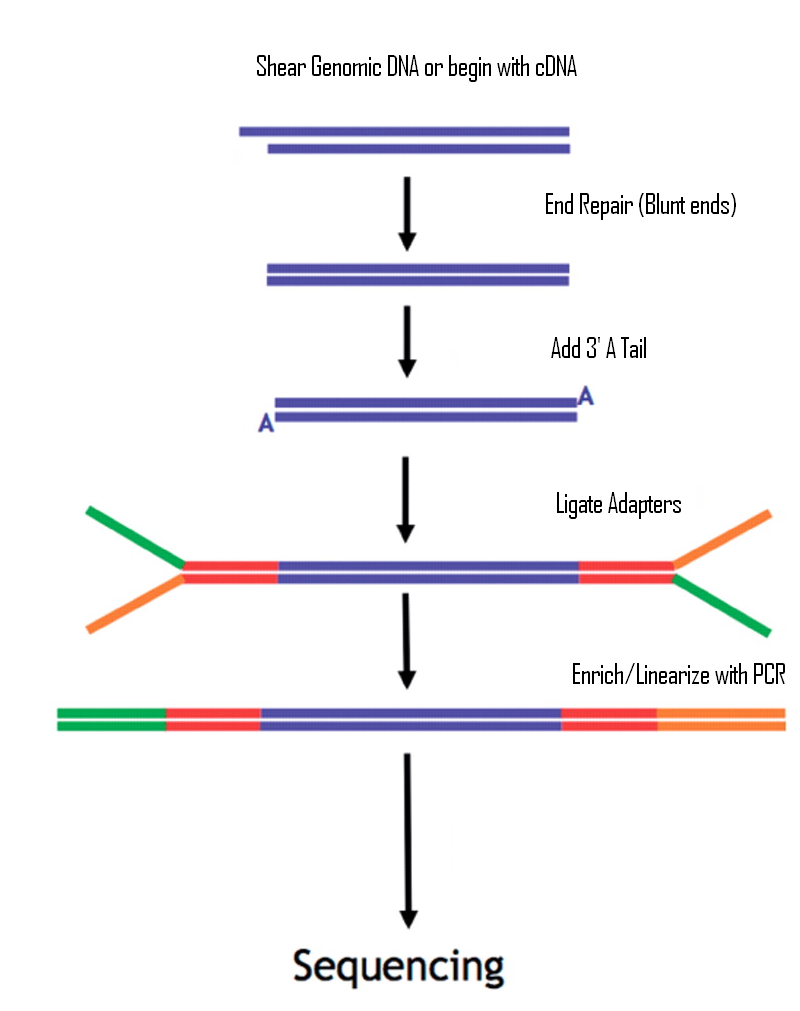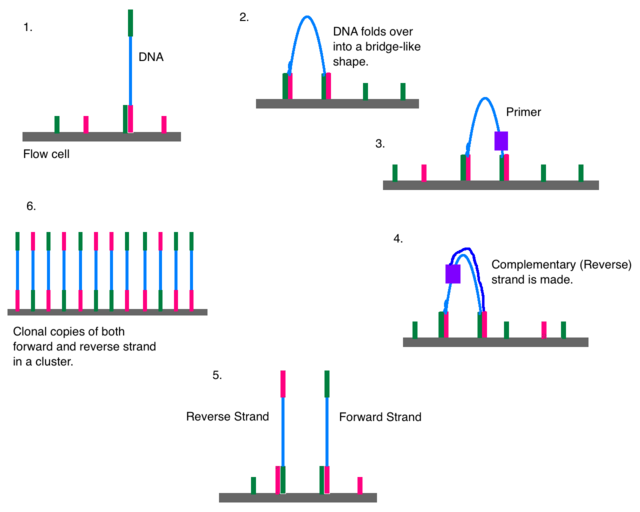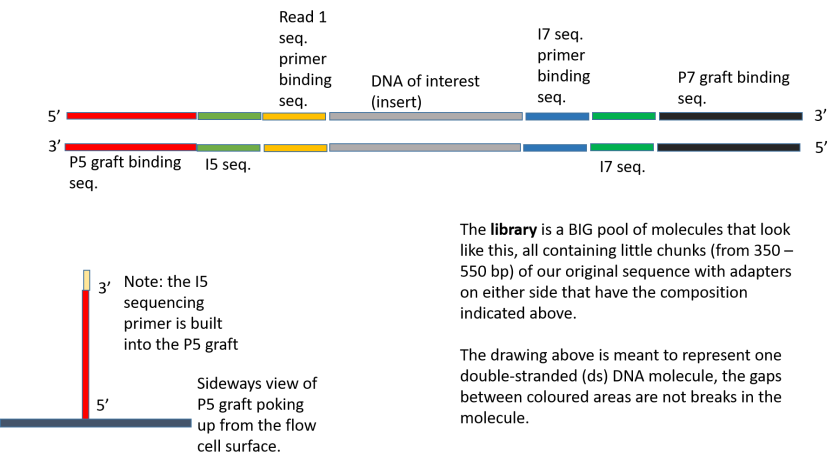
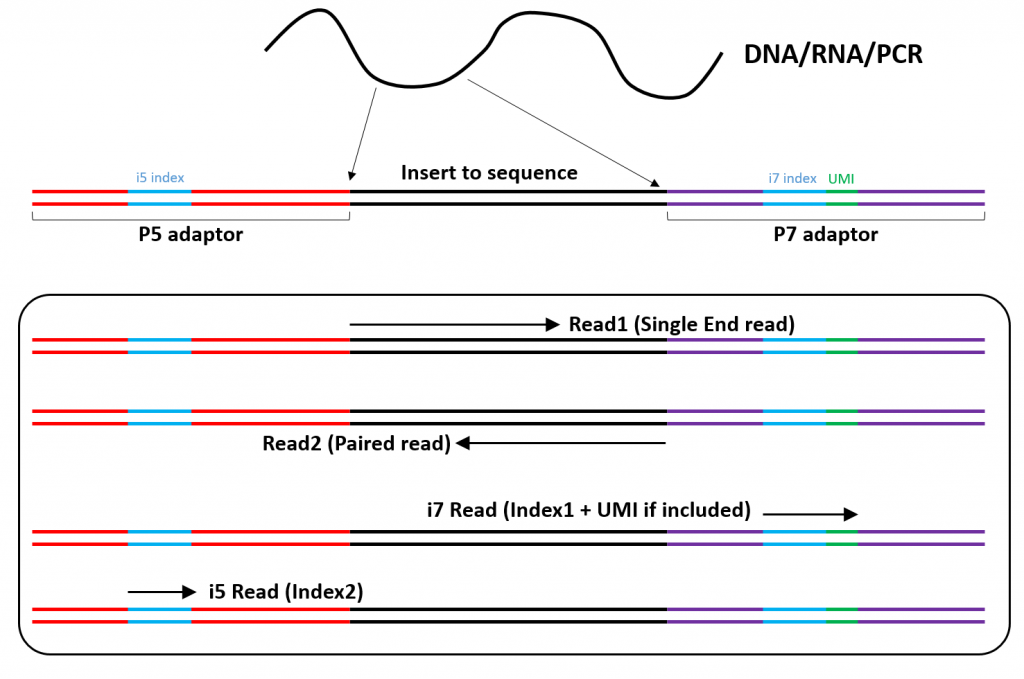
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics

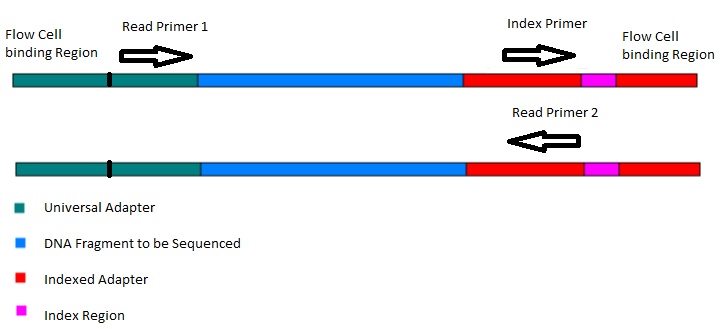
Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge
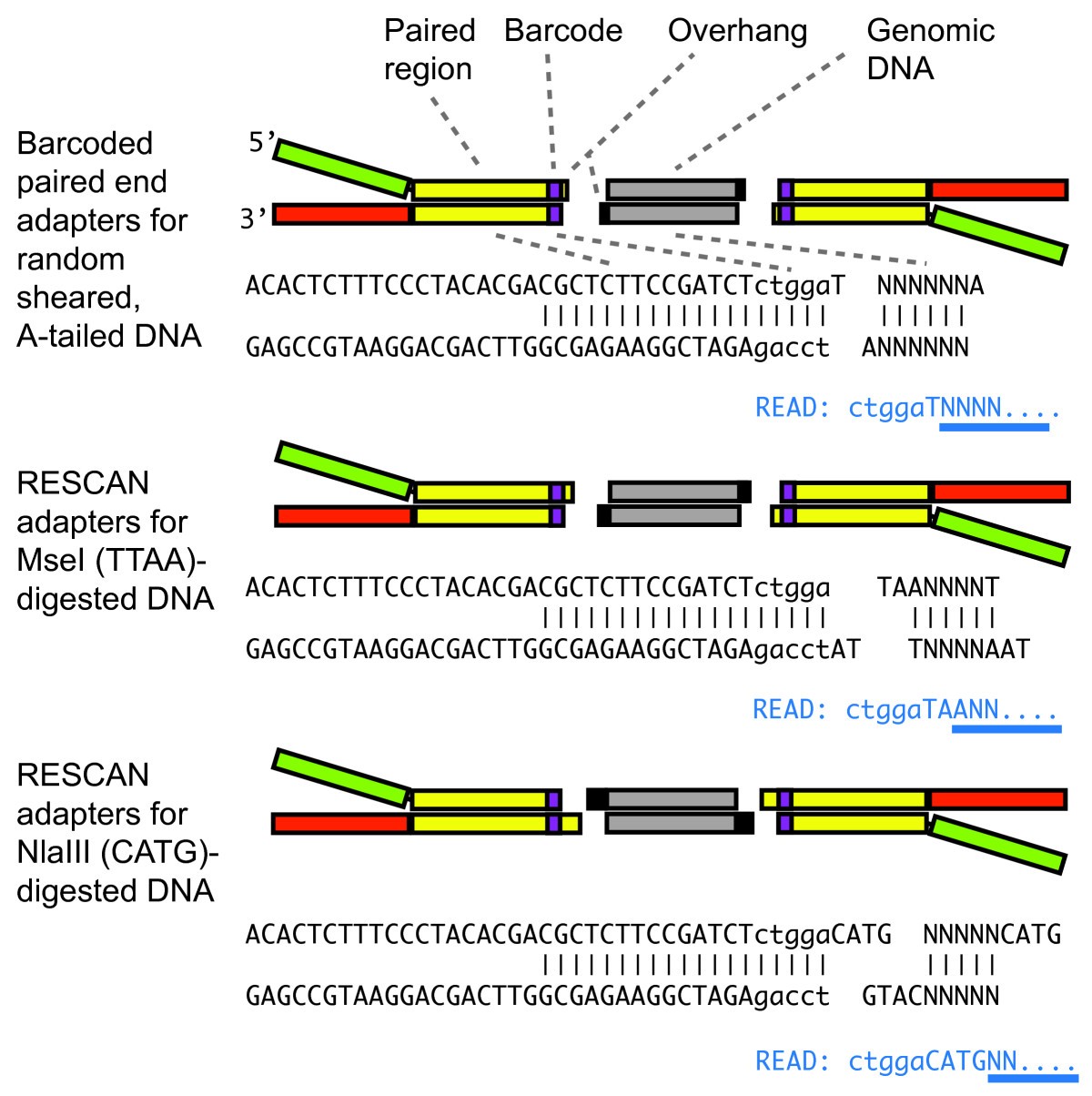
Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing | BMC Genomics | Full Text
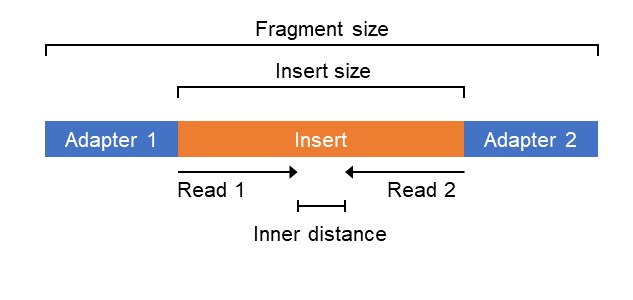
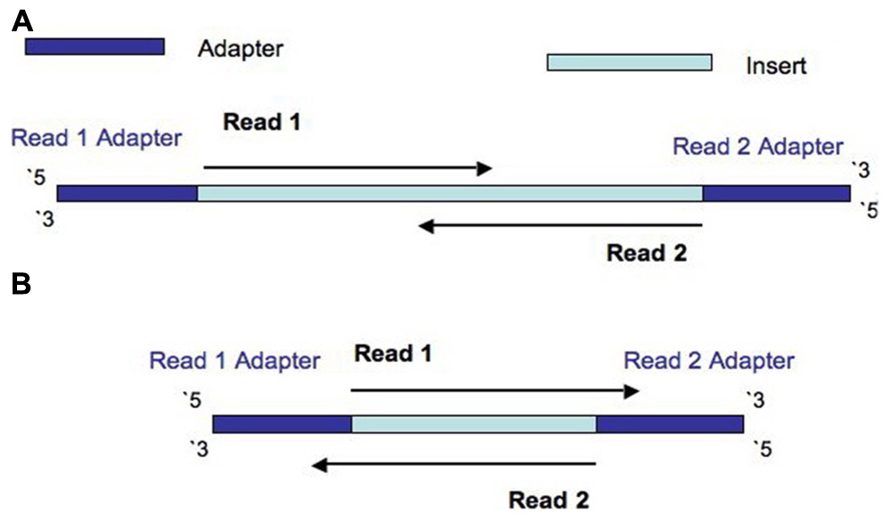
Compbio 020: Reads, fragments and inserts - what you need to know for understanding your sequencing data — Bad Grammar, Good Syntax

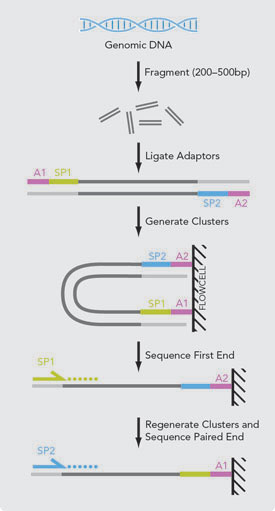


![PDF] leeHom: adaptor trimming and merging for Illumina sequencing reads | Semantic Scholar PDF] leeHom: adaptor trimming and merging for Illumina sequencing reads | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/247973613dfa3c42eb437ec9f5269202595f96af/2-Figure1-1.png)